Note
Click here to download the full example code
Comparing¶
traja allows comparing trajectories using various methods.
import traja
df = traja.generate(seed=0)
df.traja.plot()
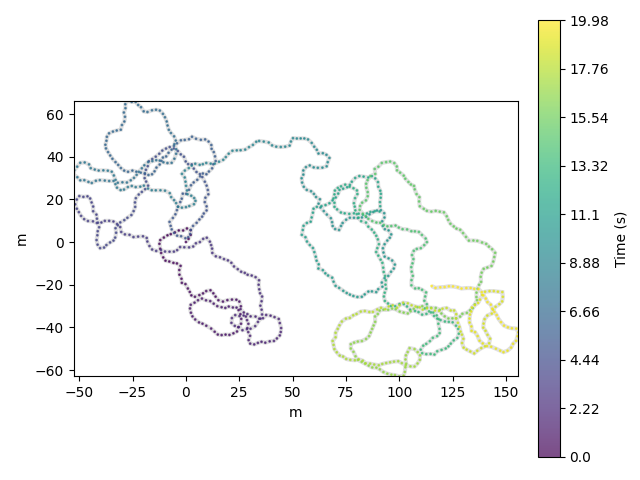
<matplotlib.collections.PathCollection object at 0x7f3c157b0110>
Fast Dynamic Time Warping of Trajectories¶
Fast dynamic time warping can be performed using fastdtw.
Source article: link.
import numpy as np
rotated = traja.rotate(df, angle=np.pi / 10)
rotated.traja.plot()
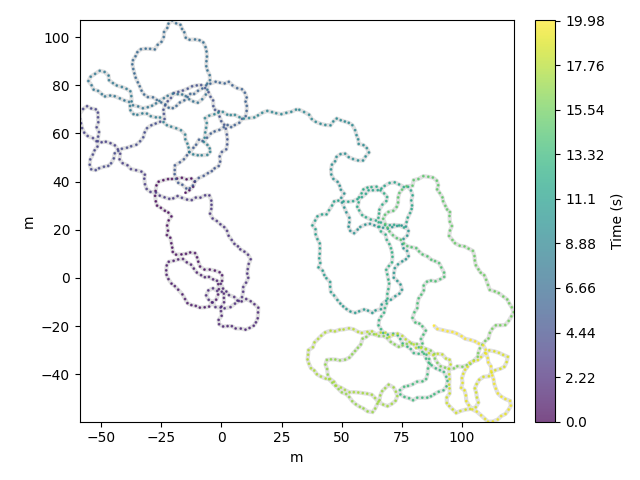
<matplotlib.collections.PathCollection object at 0x7f3b98c2d450>
Compare trajectories hierarchically¶
Hierarchical agglomerative clustering allows comparing trajectories as actograms and finding nearest neighbors. This is useful for comparing circadian rhythms, for example.
# Generate random trajectories
trjs = [traja.generate(seed=i) for i in range(20)]
# Calculate displacement
displacements = [trj.traja.calc_displacement() for trj in trjs]
traja.plot_clustermap(displacements)
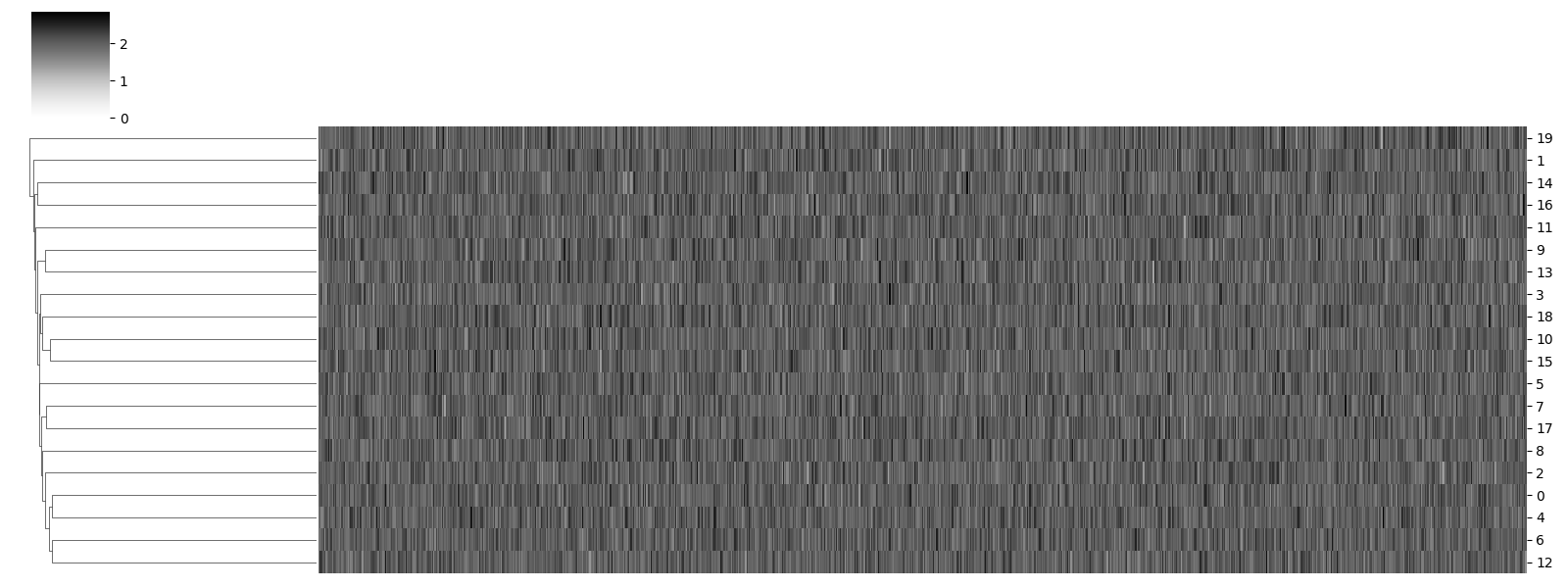
/home/docs/checkouts/readthedocs.org/user_builds/traja/envs/latest/lib/python3.7/site-packages/seaborn/matrix.py:657: UserWarning: Clustering large matrix with scipy. Installing `fastcluster` may give better performance.
warnings.warn(msg)
<seaborn.matrix.ClusterGrid object at 0x7f3ba9c31050>
Compare trajectories point-wise¶
Distance between the two trajectories is 27254.161638114736
Total running time of the script: ( 0 minutes 1.392 seconds)